Surprisingly, over 70% of the world’s antibiotics are produced using a microorganism called actinomycetes. The biosynthesis of these antibiotics often coincides with a physiological transition from primary to secondary metabolism and morphological differentiation of actinomycetes. Both developmental switches are governed by highly interconnected regulatory mechanisms at the transcriptional, translational, and post-translational levels. This study is led by Professor Byung-Kwan Cho and his research team at the KAIST Institute for the Biocentury. The work has secured a large amount of information that can be utilized in manipulating the actinomycete genome and is, therefore, expected to greatly contribute to the large-scale production of antibiotics. In addition, this study introduces the possibility of increasing the production of antibiotics by engineering the genes regulating the biosynthesis of antibiotics, which are controlled by the translation level, using synthetic biology technology.
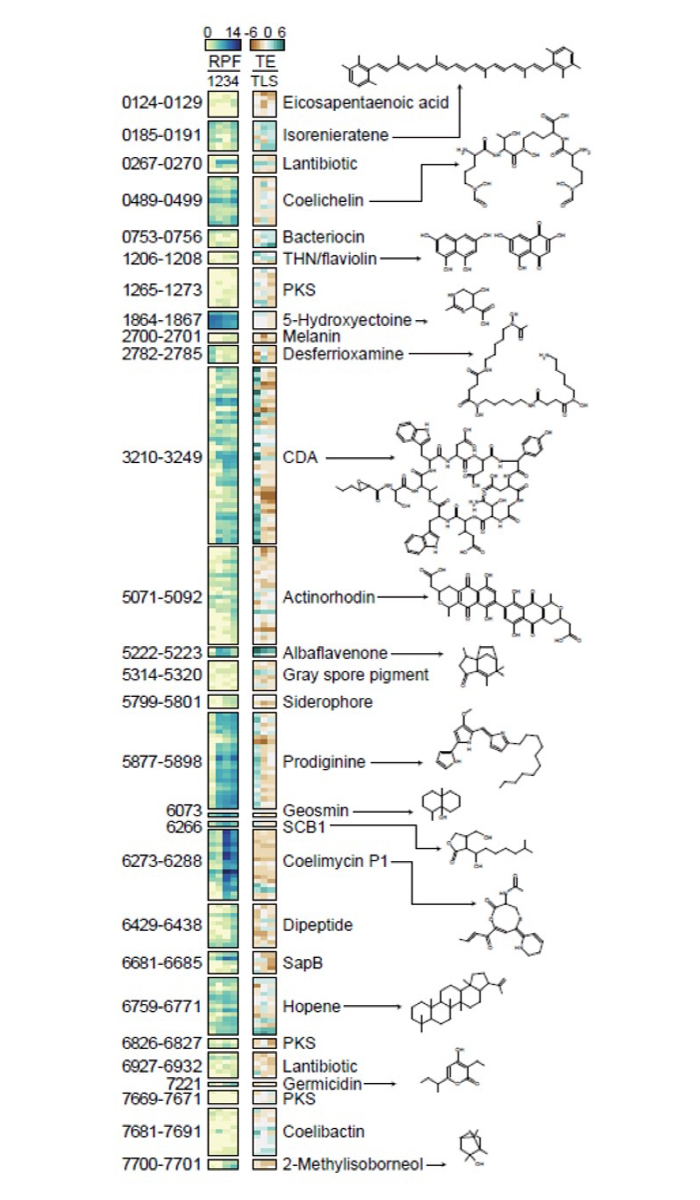
The current study was conducted to identify the overall flow of genetic information by applying three types of next-generation sequencing technology: transcription start point sequencing, RNA sequencing, and ribosome profiling. This work clarifies the gene-regulating mechanism of actinomycetes, and has secured a large amount of data, which can be utilized in manipulating the genome of actinomycetes for the mass production of various antibiotics as shown in Figure 1. Genetic structure information obtained at the genome level was used to compare gene expressions between
the levels of transcription and translation. As a result of the comparison, a phenomenon known as translational buffering was revealed. This finding shows that the translation rate at which proteins are produced from mRNA is significantly slower than the transcription rate at which mRNA is made from DNA.
The results suggest that it is necessary to consider not only the level of transcription but also the level of translation when constructing an actinomycete cell factory capable of mass production of antibiotics. The information gained on the entire genome level through the use of various next-generation sequencing techniques will be an important reference for future molecular genetic and systematic studies. Furthermore, this study also opens the possibility of increasing the production of antibiotics using synthetic biology technology by engineering the genes regulating the biosynthesis of antibiotics, which are controlled at the translation level.
Byung-Kwan Cho (Biological Sciences)
Homepage: https://www.cholab.or.kr
E-mail: bcho@kaist.ac.kr






