A joint research team comprising Dr. Gi Bae Kim, Ji Yeon Kim, Dr. Jong An Lee, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Dr. Charles J. Norsigian and Professor Bernhard O. Palsson of the Department of Bioengineering at UCSD has developed DeepECtransformer, an artificial intelligence that can predict the enzyme functions from a protein sequence, and has established a prediction system by utilizing the AI to quickly and accurately identify the enzyme function. The study was published in Nature Communications on 14 November, 2023 (Functional annotation of enzyme-encoding genes using deep learning with transformer layers, Nature Communications, 14:7370 (2023)).
Enzymes are proteins that catalyze biological reactions, and identifying the function of each enzyme is essential to understanding the various chemical reactions that exist in living organisms and the metabolic characteristics of those organisms. Enzyme Commission (EC) number is an enzyme function classification system designed by the International Union of Biochemistry and Molecular Biology. In order to understand the metabolic characteristics of various organisms, it is necessary to develop a technology that can quickly analyze enzymes and EC numbers of the enzymes present in the genome.
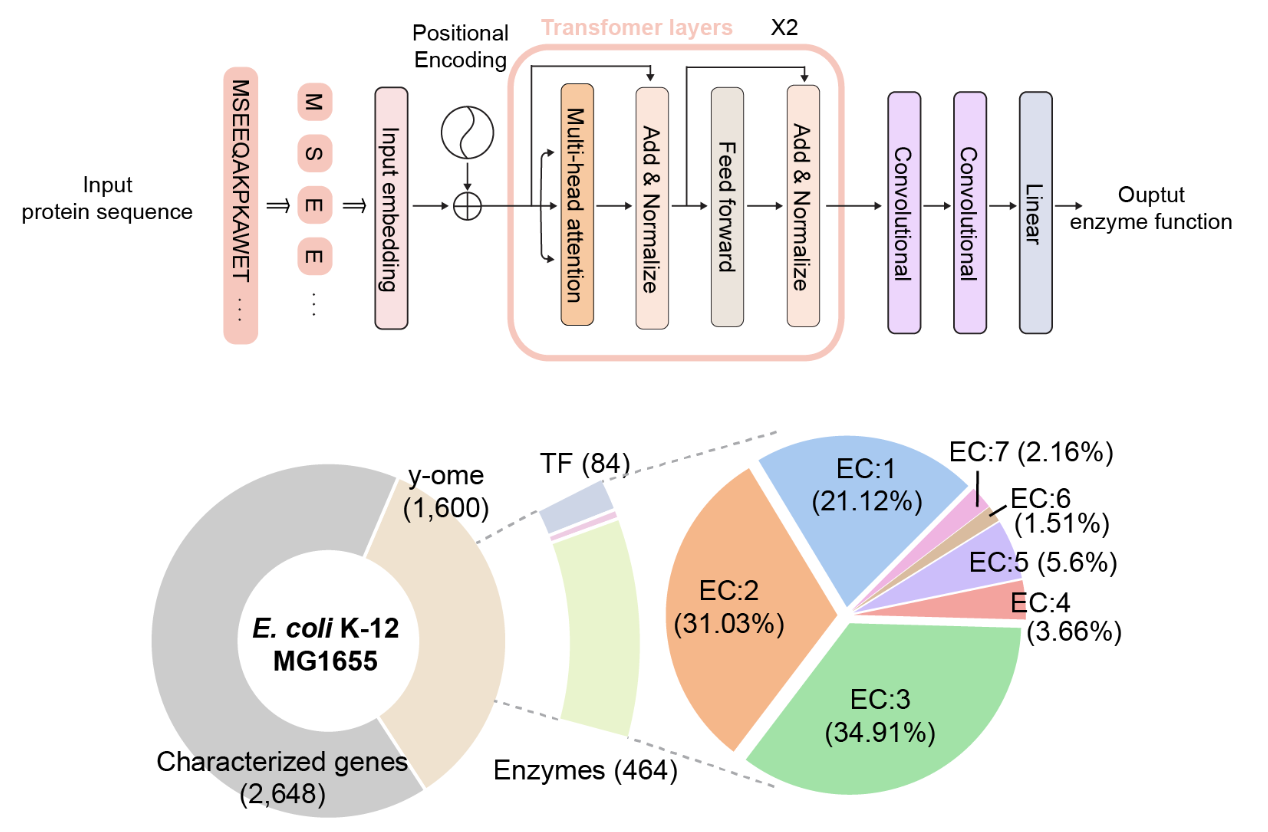
The joint team developed DeepECtransformer, an AI that utilizes deep learning and a protein homology analysis module to predict the enzyme function of a given protein sequence. To better understand the features of protein sequences, a transformer architecture, which is commonly used in natural language processing, was additionally used to extract important features about enzyme functions in the context of the entire protein sequence. This enabled the team to accurately predict the EC number of the enzyme. The developed DeepECtransformer can predict a total of 5360 EC numbers.
“By utilizing the prediction system we developed, we were able to predict the functions of enzymes that had not yet been identified and verify them experimentally,” said Dr. Gi Bae Kim, the first author of the paper. “By using DeepECtransformer to identify previously unknown enzymes in living organisms, we will be able to more accurately analyze various facets involved in the metabolic processes of organisms, such as the enzymes needed to biosynthesize various useful compounds or the enzymes needed to biodegrade plastics,” he added.
“DeepECtransformer, which quickly and accurately predicts enzyme functions, is a key technology in functional genomics, enabling us to analyze the function of entire enzymes at the systems level. Also, we will be able to use it to develop eco-friendly microbial factories based on comprehensive genome-scale metabolic models, potentially minimizing missing information of metabolism,” said Professor Sang Yup Lee.
This research was conducted with the support under the “Development of next-generation biorefinery platform technologies for leading bio-based chemicals industry project (2022M3J5A1056072)” and the “Development of platform technologies of microbial cell factories for the next-generation biorefineries project (2022M3J5A1056117)” from the National Research Foundation supported by the Korean Ministry of Science and ICT (Project Leader: Distinguished Professor Sang Yup Lee, KAIST).
Distinguished Prof. Sang Yup Lee, Dr. Gi Bae Kim Dept. of Chemical and Biomolecular Engineering, KAIS
E-mail: leesy@kaist.ac.kr
Homepage: https://mbel.kaist.ac.kr






